
Early Steppe population does not have the Y-chromosome marker that is present in a large proportion of Indians even today: Indians had migrated to Steppe, and not the other way around*.
NOTE: This is an overview counter to both the lay-press hype, which was unnecessary and unethical (please see below), and the context and setting of the methodologies used in the so-called scientific study by Reich and Co of Harvard and his compatriots. A detailed technical and scientific evidence-based (from previous published peer-reviewed work) will be published here and/ or in the appropriate scientific and academic forums. *For a deeper technical prelude (and updates) please watch Dr Priyadarshi’s current blog – The Aryan Invasion Issues.
SEPOYS OF HARVARD’S THIRD REICH GALORE
The Christian British colonialists did not rule India by having a huge army of a Christian British White soldiers, but an army made almost entirely of Indian sepoys. One would have imagined that with the 1947 transfer of power that the sepoys would have reformed themselves and converted the transfer of power from Britain to India into a real independence.
From Nehru to the academic foot soldiers, everybody became a sepoy, again. Recently, the recruitment appears to have only widened and deepened, perhaps in preparation for the 2019 elections war. Now the war is fought in the academia, in the media and the wider perception engineering arena of the PR world, the intellectual Kurukshetra. (Malhotra 2018)
Many people would criticise this author for such a polemical start for an article that is written to counter the professional work of (the so-called) scientists and academicians. That criticism is not valid as these sepoys have started behaving increasingly unethically. It has already been pointed out that the hype that the lay press created was unethical given the paper is pre-peer-review and has only been submitted to an open-peer-review platform, bioRxiv (pronounced ‘bio archive’). Of course, the sepoy authors and their Western White masters (publicly called collaborators to give a false semblance of equality) have now gone further. They have directly “leaked” their data to the lay press and in addition given open interviews where they have spouted false, or at least non-peer-reviewed, pre-submission-based conclusions. This is completely out-of-the-way from the standard ethical practice and leads one to rightfully believe that this is pure propaganda and just a hit job, not science. Is 2019 elections in their mind? Have these sepoys been knowingly bought, or inadvertently collaborating? Why stoop so low? Even one is made to wonder if Cambridge Analytica is involved? (Murali KV 2018)
CONCLUSIONS CHANGE EVEN AFTER PEER-REVIEW AND PUBLICATION
It is important to note here that the conclusions of a scientific paper is many a times changed even after it has been published. For example, a pro-Aryan-invasion “scientist” from Calcutta claimed that the phenomenon of caste-by-birth formed during the Gupta era. (Basu, Sarkar-Roy, and Majumder 2016) However, using the exact same data from the Calcutta scientist’s genetic data a reanalysis was published in the same journal altering the conclusions completely, and showing that the caste-by-birth formed only during the Islamic era. (Vadivelu 2016)
| So the gloating in the lay press, posting tweets with links to pre-peer-review copies of articles, with pre-submission pre-peer-review conclusions based on what appears to be a faulty analysis now makes one question, “Can these ‘scientists’ be even trusted with the raw data they produce, that too from very valuable ancient heritage / the nation’s biological intellectual property (IP)?” |
It also raises very interesting issues to think – how come within hours of these pre-peer-review articles being posted a whole multi-thousand worded article, with slides and elaborate graphics, gets posted on a lay press online portal and that too by an author who has no scientific background? Who gives them the data beforehand?
It is common practice for large journals to provide ‘previews’ and short write-ups (such as press notes, releases, etc.) to the lay press, before publication, of scientific papers of public interest by their respective PR desks. However, it is not known of any ethical scientist or journal to do that before peer-review, and immediately after submission, or even prior to that. Do you smell anything fishy?
NOW LET US GET TO THE SCIENCE
Note: This is based on the assumption that these “scientists” data can be trusted. It is indeed worrying after taking note of the above issues, the repeated ethical infarctions exhibited by these “scientists,” and what had been written before. (Murali KV 2018; Shukla and Venkataraman 2018; Chavda 2018)
Though this author places the validity of linguistics at a very low ranking, one should note the ridiculousness of the claims that “Indo-European” (IE) languages came from the Steppe or Central Asia. The region claimed is almost completely devoid of IE languages, but is filled with speakers of what was previously called the Altaic family. Please see the map below.
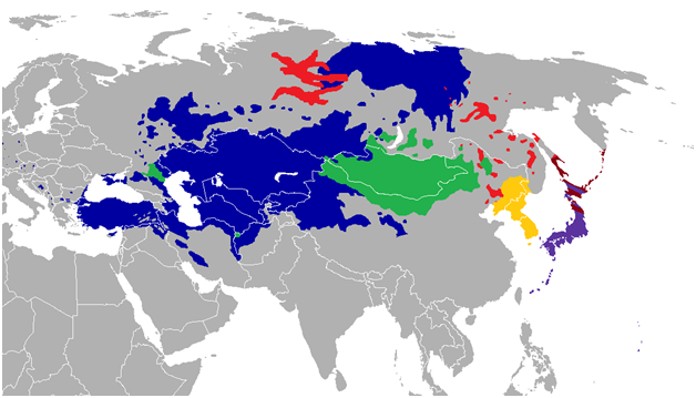
Did the Steppe people take a flight to Haryana? The coloured regions do not speak the so-called Indo-European languages
It has already been pointed out by writers in Swarajya Magazine that the Brahmins of the South India do not have any excess Steppe genetic component in their DNA in the words of the authors of the “scientific” paper themselves. In fact, by their own data and analysis, 43 percent of people who appear to have an excess of Steppe genetic component are non-Brahmins and non-Bumihars. If the Brahmins are the outsiders who brought the Indo-European languages to India from the Steppe or Central Asia, where did the Brahmins of the South India come from who have no excess Steppe DNA ancestry compared to the rest of the population? If they imported Sanskrit along with their arrival why not all Brahmins in the North, South, East, Central and West India admittedly have the excess Steppe contribution, but only some.
| Clearly this belies that there is an association, and causation, between Sanskrit and so-called Steppe ancestry in their own flawed analysis of their own data. These kind of absurdities, if qualified as good science (because it comes from Harvard and team), then anything goes as science to say the least. It is likely they cannot differentiate correlation vs causation. |
By these authors own criteria and classification of the data, as revealed in one tiny part of the pre-peer-review article, namely Figure 2E, the early Steppe_EMBA samples have no R or R1a Y-chromosome DNA markers. But they suddenly appear in later samples, Steppe_MLBA and Steppe_LBA, indicating these arrive to the Steppe from somewhere else. This clearly contradicts their own fancy hypotheses. Previously it is known that the predominant Indian R1a, R1a-Z93, is different from the European R1a. So we also need to know the type of R1a these Steppe samples appear to have in the latter periods. (Priyadarshi 2014, 2011)
About a week ago, an Indian scientist who has recently (2016–17) been awarded a visiting fellowship at the Harvard Medical School (where the new age race-theorist Third Reich is a faculty), and with access to ancient Indus Valley Civilisation (IVC) bone samples with the ancient DNA in them, has claimed the following in an interview with the Open Magazine before the data has been submitted, peer-reviewed and published, “While Rai and Shinde are tight-lipped about what the DNA samples extracted from the Indus Valley remains show, Rai does reveal that the R1a genetic marker is missing in the sample. This is a significant revelation. R1a is believed to have originated sometime between 22,000 and 25,000 years ago.”
During the last ice age (about 18,000 years ago), the northern latitudes were all severely cold and bone dry and humans went extinct. This ice age (last glacial maximum, LGM) ended about 13,000 years ago.
| So if R1a originated in 22,000 and 25,000 years ago, and has survived the ice age, it could have survived only in the tropical Africa, India and South East Asia. It is well known that the R1a lineages are found only in India and Eurasia north and west of India. It is present in East Africa from migration from India. So, if these scientists widen their horizons, look beyond propaganda and study other specialities of science, they will see the facts right. (Priyadarshi 2014, 2011) |
If it is missing in the IVC samples and is also missing in early Steppe samples, it must have got to both the places from somewhere else. Kindly note that this is assuming that the current samples from IVC is not skewed by sampling. What is sampling and how it is relevant will be explained below. We know from previously published peer-reviewed scientific research that the South Indian populations are (both the caste groups and long-isolated tribes) rich in R1a Y-chromosome DNA markers. (Priyadarshi 2014, 2011)
This brings a huge evidence-based hypothesis to the table – is it the South Indians who migrated to IVC and the Steppe about 3500–4000 years ago?* This will explain the relatively smaller proportion of the so-called South Indian DNA (the hypothesised, Ancestral South Indian [ASI] and Ancient Ancestral South Indian [AASI]) in Northwest India, about 20 per cent depending on the study and source data. (Kivisild et al. 2003; Sharma et al. 2009)
Perhaps, to deliberately hide the South Indian ancestry from Steppe and elsewhere, did they use the Andamanese population’s (Onge) DNA as the control? The Andamanese population is known to have separated from India 40,000 years ago without any admixture thereafter. The further away in time and geography a control population is, the DNA analysis will more likely show no or only a very distant relationship.
With over 90 authors involved in the paper, if so many defects are identifiable even before the complete data is available, which is usually available as supplementary data on publication, should not one consider a deliberate attempt at painting a certain picture (a.k.a. propaganda)? Remember, all this is assuming that the data itself has not been tampered with. Usually, one gives the benefit of doubt to the scientists, but these “scientists” as they say in the legal circles have “not come with clean hands.” (Baker 2016)
More skeletons might come out of the closet if the data (assuming they are reliable) are reanalysed. For now, let us assume the best, but keep our eyes and ears open.
To be continued…
Note:
1. The views expressed here are those of the author and do not necessarily represent or reflect the views of PGurus.
References:
- Baker, Monya. 2016. “1,500 Scientists Lift the Lid on Reproducibility.” Nature 533 (7604): 452–54.
- Basu, Analabha, Neeta Sarkar-Roy, and Partha P. Majumder. 2016. “Genomic Reconstruction of the History of Extant Populations of India Reveals Five Distinct Ancestral Components and a Complex Structure.” Proceedings of the National Academy of Sciences of the United States of America 113 (6): 1594–99.
- Chavda, Abhijit. 2018. “What Reich’s Study Says And Doesn’t About How Indians Came To Be.” Accessed April 29. https://swarajyamag.com/ideas/what-reichs-study-says-and-doesnt-about-how-indians-came-to-be.
- Danino, Michel. 2010. The Lost River: On the Trail of the Sarasvatī. Penguin Books India.
- Kivisild, T., S. Rootsi, M. Metspalu, S. Mastana, K. Kaldma, J. Parik, E. Metspalu, et al. 2003. “The Genetic Heritage of the Earliest Settlers Persists Both in Indian Tribal and Caste Populations.” American Journal of Human Genetics 72 (2): 313–32.
- Malhotra, Rajiv. 2018. “Introduction to Rajiv Malhotra’s Vocabulary – Infinity Foundation.” Infinity Foundation. Accessed April 29. https://infinityfoundation.com/course/rajiv-malhotra-vocabulary/.
- Murali KV. 2018. “There Are Lies, Damned Lies and (Harvard’s ‘Third’ Reich and Co’s) Statistics – PGurus.” PGurus. April 29. https://www.pgurus.com/there-are-lies-damned-lies-and-harvards-third-reich-and-cos-statistics/.
- Priyadarshi, Premendra. 2011. The First Civilization of the World. Siddharth Publications.
- ———. 2014. In Quest of the Dates of the Vedas: Comprehensive Study of the Vedic and the Indo-European Flora, Fauna and Climate in Light of the Information Emerging from the Disciplines of Archaeolo. Author Solutions.
- Sharma, Swarkar, Ekta Rai, Prithviraj Sharma, Mamata Jena, Shweta Singh, Katayoon Darvishi, Audesh K. Bhat, A. J. S. Bhanwer, Pramod Kumar Tiwari, and Rameshwar N. K. Bamezai. 2009. “The Indian Origin of Paternal Haplogroup R1a1* Substantiates the Autochthonous Origin of Brahmins and the Caste System.” Journal of Human Genetics 54 (1): 47–55.
- Shukla, Aseem, and Swaminathan Venkataraman. 2018. “Why The Latest Genetic Study Does Not Rewrite India’s History.” Accessed April 23. https://swarajyamag.com/ideas/why-the-latest-genetic-study-does-not-rewrite-indias-history.
- Vadivelu, Murali K. 2016. “Emergence of Sociocultural Norms Restricting Intermarriage in Large Social Strata (endogamy) Coincides with Foreign Invasions of India.” Proceedings of the National Academy of Sciences of the United States of America 113 (16): E2215–17.
- Priyanka Vadra continues the illegal abuse of Gandhi’s name and legacy - March 19, 2019
- Is secularism the other side of genocide and is it thus unconstitutional? - November 10, 2018
- Is Yogi’s government inadvertently putting the Kumbh Mela at subversive risk by Breaking India forces at Harvard et al? - October 17, 2018










[…] Murali KV (2018) Early Steppe vs Middle to Late Steppe Early Steppe population does not have the Y-chromosome marker that is present in a large proportion of Indians even today. PGurus.com. Available at: https://www.pgurus.com/early-steppe-vs-middle-to-late-steppe/. […]
Great article. We need more such informative writing to put western biases in their place.
All BS.
This link will provide data about DNAs of the various European countries.
http://www.khazaria.com/genetics/
[…] is the concluding part of the series Early Steppe vs Middle to Late Steppe in which the author disputes the theory of David Reich and his team at Harvard on the […]
Regarding the altaic languages,is it possible that it spread with the mongol expansion and conquest during the last millennium?
Good question, but a convoluted possibility in more than a couple of ways. Apparently, many now believe Turkic group and the Mongolic group are different families and should not be considered one. The older so-called proto-Turkic and proto-Mongolic languages are apparently less similar than modern Turkic and Mongolic. So the increased similarity in modern Turkic and Mongolic languages could be possibly attributed to Mongol expansion and there appears no replacement by these languages of any other languages, including IE-related ones.
In any case, neither language groups (Turkic or Mongolic) are IE per se, according to the linguists (which the author barely considers a science, please see the previous article for the author’s scientific opinion on it).
There are plenty of more robust genetic, climatological, archaeological, multi-species, multi-disciplinary evidence from over 10 fields – https://www.pgurus.com/there-are-lies-damned-lies-and-harvards-third-reich-and-cos-statistics/
These so-called “scientists” of the Reich camp are just ignorant or deliberately ignoring those evidence.
Just realized that the author happens to be the same person who published the re-analysis to the Basu et al endogamy study :).
“This brings a huge evidence-based hypothesis to the table – is it the South Indians who migrated to IVC and the Steppe about 3500–4000 years ago” — While south indian people do have R1a1, they mostly have derived branch R1a-Z93 which you already know. The chances of south indian folks migrating to steppe around 2000 BC bringing R1a with them doesn’t make much sense considering R1a has been found in Eastern Europe in a skeleton supposedly dated to around 5000 BC. So any migration from South India should supposedly happen earlier if one considers them as the vector of R1a .
“the early Steppe_EMBA samples have no R or R1a Y-chromosome DNA markers” — You might want to make a correction here. The early steppe folks while don’t have R1a lineage but they do other downstream lineage of R ( that is R1b).
Good question. R1b cannot become R and then R1a. So that does not prove anything. Even if R and R1a are were to be found in the earlier samples in the Steppe and around, given the date of origin of R lineages before the LGM (climatological considerations), it only could have arrived there from somewhere else where they had survived during the LGM in meaningful numbers.
Hey M,
The question is not about whole R lineage (anyways R* has been found in an skeleton in siberia dated to around 22000 BC in Russia .https://en.m.wikipedia.org/wiki/Mal%27ta–Buret%27_culture). The question is about a downstream R1a lineage ,R1a-M417 which is relatively recent and happens to be the parent of R1a-Z93 (Asian) and R1a-Z282(European) and which most likely corresponds with copper age/bronze age movements and possibly with so-called Indo-european expansions. Arrival from South Asia some 12,000 -15000 years ago won’t have much significance as there is no evidence of any language , chariot/horse stuff or even agriculture that long ago . It’s only in the vicinity of 6000-4000 years ago that would make sense in the context of Indo-european expansions.
Not only R*, but many such lineages traveled long distances much before 20,000 BP. Two such examples are C and D Y-DNAs reaching Australia and Papua New Guinea about 50,000 years back.
But unfortunately the harsh LGM peak about 20,000-18,000 BP killed the siberian and north European them all.
Another great mass killer was about 13,000-11,500 BC freeze. And a third mass killer was 8.2 kilo-year effect. It is impossible for R* to have survived these climatic catastrophes for Siberia.
By the way, there is evidence that R* possibly originated from P1 in Southeast Asia or in South Asia. From there its branch R1a had reached Bhutan also before splitting between R1a and R1b. (Hallas 2014: page 666). These data are more relevant for Holocene and not the Ice Age data from Siberia, which at the most tell us the high spirits of our Palaeolithic ancestors. And that indicates, they too–the people of 20,000 or 50,000 ybp–did speak some language.
So do you also mean to say that the Y-DNA H found in Early Neolithic Europe, the Y-DNA F* also found in early Neolithic Europe etc did not migrate from India?
Prem sir, I thought the Y-DNA found in early neolithic Europe belongs to H2 (P96) Branch which is not found in South Asia. Good point about F* .
Thanks for the Hallast information, will read the paper.
If one gets neolithic and copper age remains from south asia , a lot of things might become clear
H2 is not found in India today, but does that mean it originated in Israel or Europe? Do you mean H2 is born from J or K or G, or directly from A or B?
Being H2, it has to be born from H, and thus is a brother of H1. By the way, H1 is also found in the modern population of Ukraine, Serbia and Macedonia. Don’t use this evidence to claim that H1 too arrived to India from the steppe or southeast Europe.
Prem sir, I thought the very small % of H1 found in Ukraine,Serbia is due to Romani people. Good point about H2 but i would assume that the split between H1 and H2 would have happened some 20,000 years ago (or even older) . Maybe be the split took place in North-west india/Pakistan between H1/H2 with H2 moving out but with arrival of new haplogroups like J2,L ,H1 got replaced upto an extent in North west India !!! 😛
The figures I cite are for general white population of Europe. As per census, the Romani population is only 45,000 out of 45 million population of Ukraine. That makes Romani 0.1% of Ukrainian population. On the other hand H1 is 1.1% of Ukrainian population. So you mean Romanian each male reproduced into White Ukrainian population 10 male children. And this also assumes that they were all H1a!!! This is how you people create weak argument which collapses the test of scrutiny.
http://www.khazaria.com/genetics/ukrainians.html
See link.
What a joke! H in Europe is because of a minuscule amount of Romanis. However, neither South Indians did migrate to North India and further nor Indians did migrate to Central Asia, Middle East and further to Europe. What a hypocritical stand! No wonder Reich and company have been called Third Reich…
Prem sir,
Regarding romani/Gypsy population in Europe, https://en.m.wikipedia.org/wiki/Romani_people_in_Ukraine
Quoting a line – ” In reality, by the preliminary estimates of communication within our nation, only the East of Ukraine has approximately 150 thousand Romani nationals, and we are sure that the Romani population on the territory of Ukraine reaches more than 400 thousand people. ”
More stuff here
http://www.minelres.lv/reports/ukraine/PDF_Roma_of_Ukraine_eng.pdf
Now regarding Ukraine https://en.m.wikipedia.org/wiki/World_War_II_casualties , the data estimate that close to 16.3% people died in Ukraine. Such a scenario where large number of males die can cause decrease in some Y-lineages and subsequent founder-effect in some lineages during repopulation can cause increase in other Y-lineages.
“However, neither South Indians did migrate to North India and further nor Indians did migrate to Central Asia, Middle East and further to Europe. What a hypocritical stand! ” –> Hey M, we know that there were people moving from south india. South Indian specific mtDNAs were found in Mesopotamia (we know south india had extensive trade relations vis sea route so finding indian specific lineages in Mesopotamia,east africa shouldn’t surprise anyone. Like wise we have Near East specific Y-HG J2 found in large numbers in Western belt of India). However I am not sure of any large scale migration from south india in the past 7000 years though . The admixture plot doesn’t reveal it though (However, very old remains found in siberia like Ma’lta Buret boy and Ust-Ishim do show some autosomal affinity with present day south indians :/ ).
When i said “due to romani people” I meant males from those marrying with the girls of local population and inducted into the local population.
Another set of scientists involved with Riech has published another hit job interview viz a viz IVC genome markers before even publishing the paper in bio archive. http://www.caravanmagazine.in/vantage/indus-valley-genetic-contribution-steppes-rakhigarhi
Dr Roy’s job is pukka (confirmed) in Harvard now.